Today I will introduce you to the methods for RNA and protein analysis at massive scale - **RNA sequencing (RNA-seq)** and **tandem mass tag mass spectrometry (TMT-MS)**. These two methods provide as an output enormously large amount of data that needs to be properly analysed in order to obtain the biological meaning of phenomena you're investigating. <center>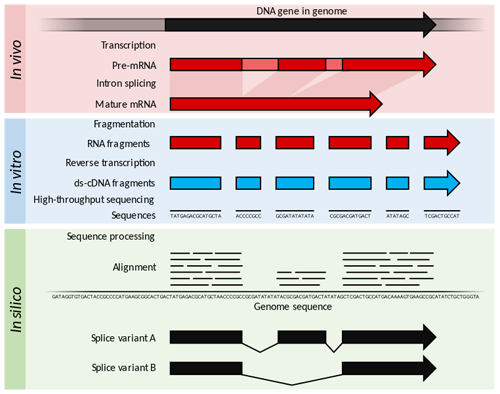</center> <center>[Summary of RNA-Seq](https://en.wikipedia.org/wiki/File:Summary_of_RNA-Seq.svg) by Thomas Shafee, [CC BY 4.0](https://creativecommons.org/licenses/by/4.0/deed.en)</center> ### Trancriptome and central dogma of molecular biology _______ Before we dive into the description of RNA sequencing methodology, first we need to explain certain basics of molecular biology. <div class=pull-left> 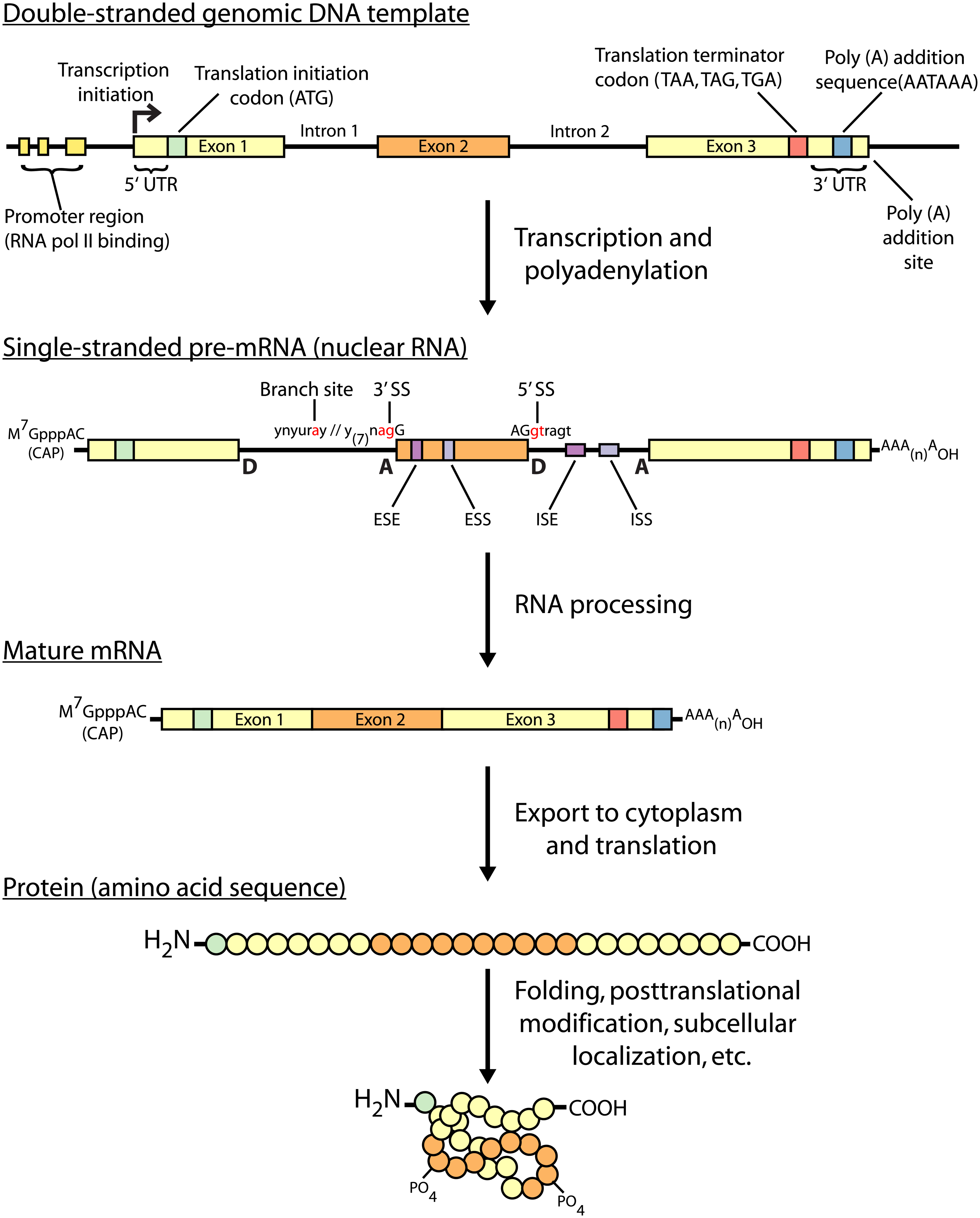 <center> <sup>[Central dogma of molecular biology](http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004393), taken from [1], [CC BY 4.0](https://creativecommons.org/licenses/by/4.0/deed.en)</sup> </center> </div> <div class=pull-right> According to the **central dogma of molecular biology**, genetic information contained within the DNA molecule is *transcribed* into the **pre-mRNA**, which is further processed and *spliced* into the **mature mRNA**. Mature mRNA is then exported from nucleus to cytoplasm, where is being *translated* into protein product, which is modified, folded into its final conformation and transported into the right cellular compartment. Now that we know what is mRNA, we can define **transcriptome**, which represents a complete set of mRNA transcripts contain in one cell. </div> <br><br><br><br> <br><br><br><br><br><br><br><br><br><br><br> <br><br><br><br><br> ### RNA sequencing _______ RNA sequencing allows RNA analysis through direct determination of cDNA sequence, and it's based on high-throughput next-generation DNA sequencing (NGS) technologies. <div class=pull-right> 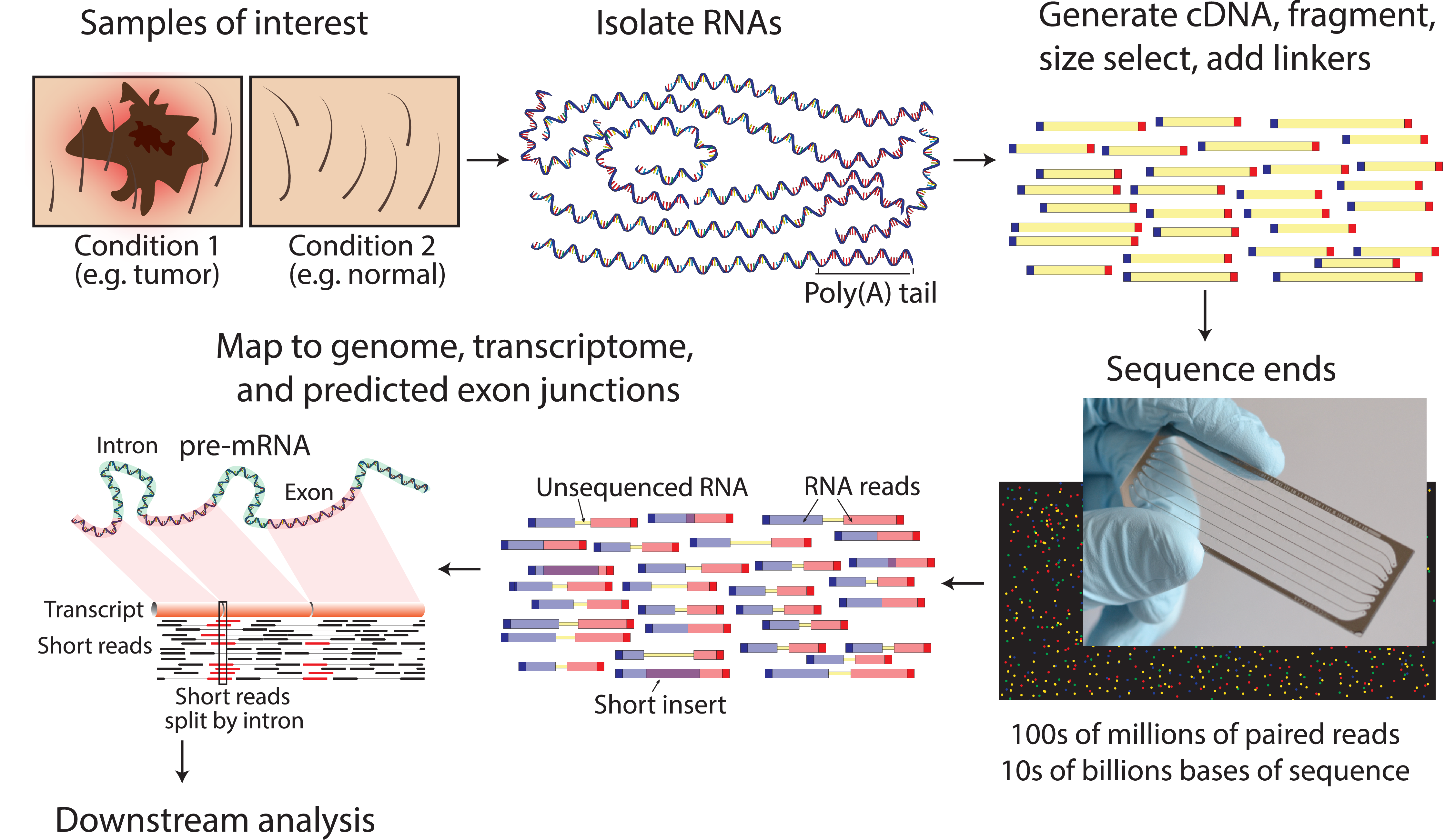 <center> <sup>[RNA-seq procedure](http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004393), taken from [1], [CC BY 4.0](https://creativecommons.org/licenses/by/4.0/deed.en)</sup> </center> </div> <div class=pull-left> Briefly, mRNA is isolated from samples (in our case, [transgenic cell line I personally made](https://steemit.com/steemstem/@scienceangel/lab-diaries-2-making-my-own-transgenic-cell-line)), and using the process of *reverse transcription* mRNA is transcribed into cDNA (complementary DNA). This collection of cDNA is called **cDNA library** and contains cDNA fragments with **adaptors** attached on both ends. After that, each molecule is being sequenced and millions of short reads are produced. </div> <br><br><br><br> <br><br><br><br><br><br><br><br><br><br> After the sequencing process is finished, filtering of reads and adapter trimming are performed, followed by *de novo* assembly of transcripts (in case that reference genome is not available) or mapping (alignment) of sequencing reads to reference genome or transcriptome. Beside all of the above steps, many other "adjustments" of the raw data are necessarily performed, until the expression values are finally obtained: 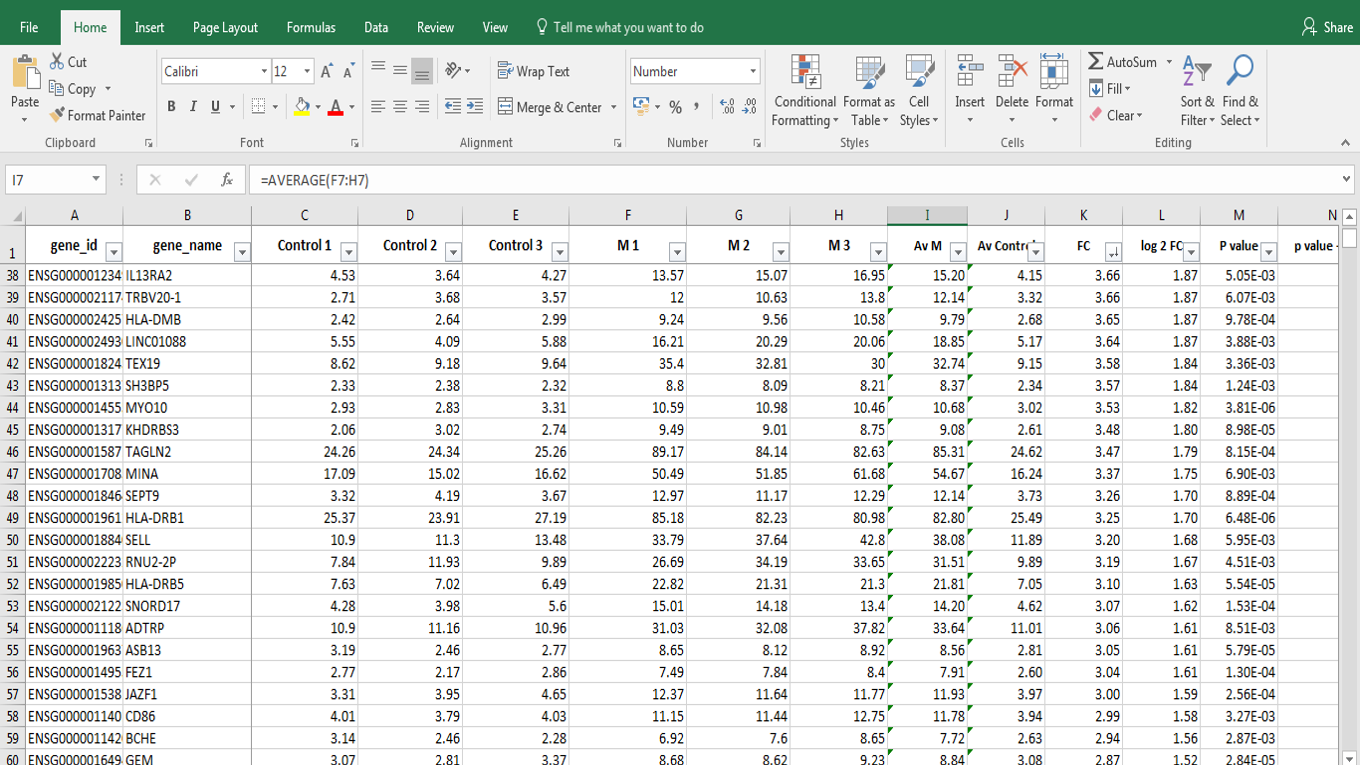 *Screenshot of my RNA-seq data* In these data I'm analyzing expression values are represented as **TPM (Transcripts Per Kilobase Million)**. This means that these data are normalized to gene length and sequencing depth, which allows us to compare normalized reads between different samples. In my RNA-seq data I have 9616 different genes for which I have to analyze expression values and compare them to control values! In order to filter data out, I perform standard Student's t-test to obtain p values for each gene, which will help me discard data that are not statistically significant. After filtering for p < 0.05, I got **5102** statistically significant, differentially expressed genes left - still sounds like a lot, don't you think? :) ### Proteome analysis by TMT-MS _______ So we have determined the expression values for mRNA extracted from our model system, but is that enough to draw conclusions on what's going on in our cells over-expressing our gene of interest? Well, not exactly, because if you remember the central dogma, mRNA is translated into protein in cells - so it would be useful to know the amount of each protein product as well. One of the ways to quantify proteins within the sample is to use **tandem mass tag mass spectrometry (TMT-MS)**. <div class=pull-left> 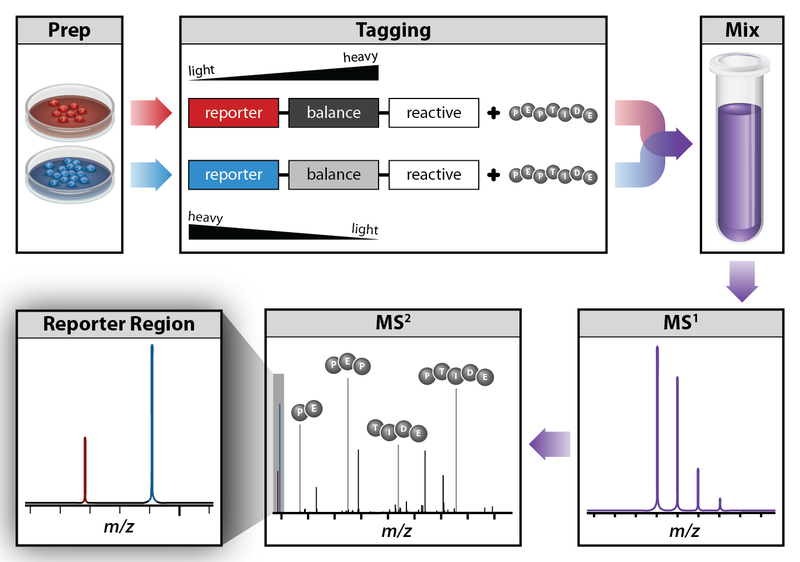 <center> <sup>[A schematic of isobaric labeling](https://en.wikipedia.org/wiki/File:Isobaric_labeling.png), by A.J. Bureta, [CC BY-SA 3.0](https://creativecommons.org/licenses/by-sa/3.0/)</sup> </center> </div> <div class=pull-right> Tandem mass tags (TMT or TMTs) represent chemical labels that are used for quantification and identification of biological macromolecules (eg. proteins). TMTs belong to isobaric mass tags, meaning that those chemical groups have the same mass. The method is based on pairing lighter with heavier regions of tags, in such way that the entire tag when attached to the peptide adds the same mass shift, which enables detection of the amount of each peptide. </div> <br><br><br><br> <br><br><br><br><br><br><br><br><br><br><br><br> What we get as output data after raw data analysis is normalized relative abundance (%) of each protein: 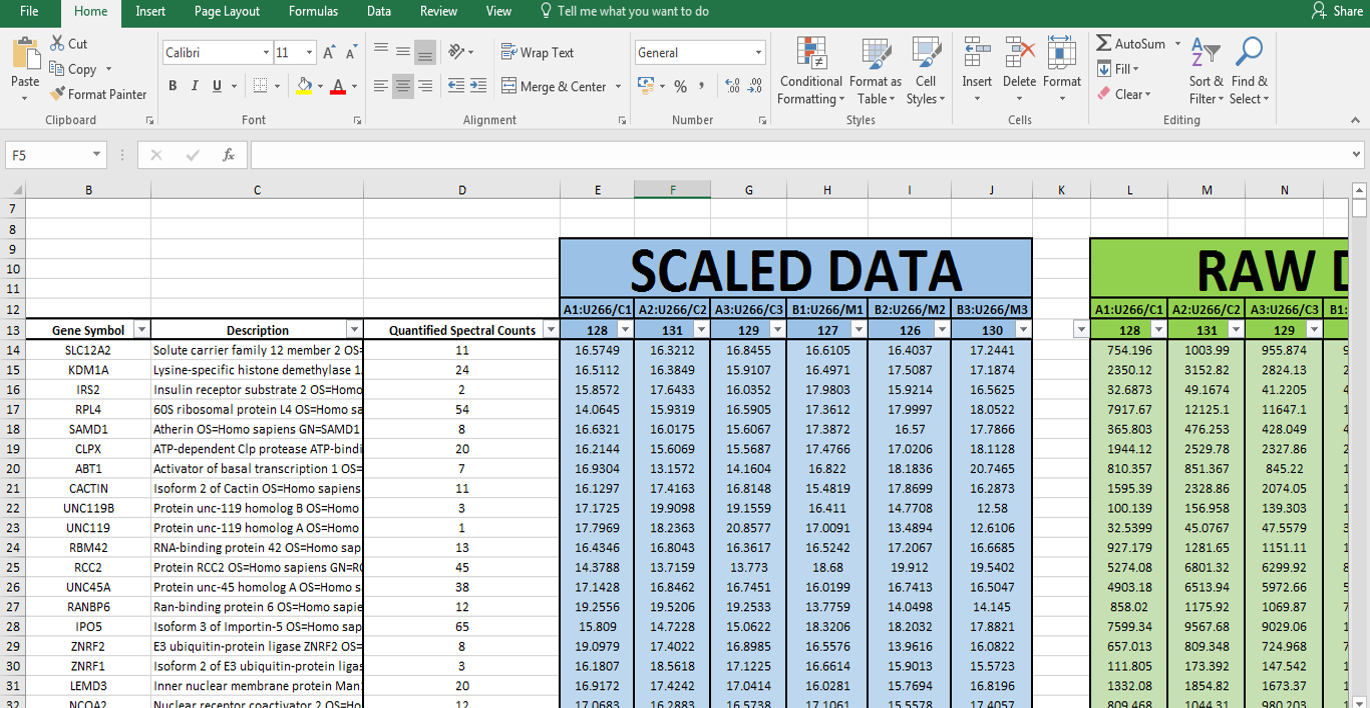 *Screenshot of my TMT data* Initially, I got 7260 detected proteins, and after I performed Student's t-test and obtain p values for each protein (same as with RNA-seq data), I ended up with **2844** statistically significant (p < 0.05), differentially expressed proteins. ______________ In the next Lab Diaries post, I will explain which method I use for analysis of such large-scale RNA-seq and TMT-MS data, and how do we obtain biological meaning from such enormous amount of data. Until then, relax and keep steemSTEM! ;) ___________ Literature [1] [Griffith, M., Walker, J. R., Spies, N. C., Ainscough, B. J., & Griffith, O. L. (2015). Informatics for RNA sequencing: a web resource for analysis on the cloud. PLoS computational biology, 11(8), e1004393.](http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004393) [2] [Ozsolak, F., & Milos, P. M. (2011). RNA sequencing: advances, challenges and opportunities. Nature reviews genetics, 12(2), 87.](https://www.nature.com/articles/nrg2934) [3] [Wang, Z., Gerstein, M., & Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews genetics, 10(1), 57.](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2949280/) [4] [Kukurba, K. R., & Montgomery, S. B. (2015). RNA sequencing and analysis. Cold Spring Harbor protocols, 2015(11), pdb-top084970.](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4863231/) [5] [RPKM, FPKM and TPM, clearly explained](http://www.rna-seqblog.com/rpkm-fpkm-and-tpm-clearly-explained/) [6] [Zhang, L., & Elias, J. E. (2017). Relative protein quantification using tandem mass tag mass spectrometry. Proteomics: Methods and Protocols, 185-198.](https://www.ncbi.nlm.nih.gov/pubmed/28188531) [7] [Tandem mass tag](https://en.wikipedia.org/wiki/Tandem_mass_tag) [8] [Isobaric labeling](https://en.wikipedia.org/wiki/Isobaric_labeling) -------------- #### For more scientific-related content check [steemSTEM](https://steemit.com/trending/steemstem). Follow me if you like my posts and want to read some more ;) If you have any thoughts/suggestions fell free to leave a comment! <center> </center><center> Image credits: [Jason Brooks](https://www.jason-brooks.com/) and [Prof. Sharona Even-Ram](https://medicine.ekmd.huji.ac.il/En/Publications/ResearchersPages/pages/sharonaev.aspx)</center>
| author | scienceangel |
|---|---|
| permlink | lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms |
| category | steemstem |
| json_metadata | {"tags":["steemstem","steemeducation","science","biology","life"],"image":["https://steemitimages.com/DQmb3BbGfvtYoQ4oVYTYvX8H8WojtVtk5DGVp1ukT6nDqjr/762px-Summary_of_RNA-Seq.svg%20small.png","https://steemitimages.com/DQmdGoJTSfLC1D53LfEehXLrKbji2HYrWFYScy9BmQA9Wo5/journal.pcbi.1004393.g001.PNG","https://steemitimages.com/DQmPEAjcSxbKTJPXszsfGxzxrNHWjMuqDEhMYMkgi9vu8Qc/journal.pcbi.1004393.g002.PNG","https://steemitimages.com/DQmTY2tRMJWdmXLZhv3rPALB5qVouBUkj9cRWHbrNHKyahm/rna%20seq.png","https://steemitimages.com/DQmNP8hrenAbZQvgfBjuM4WuThThpwtgSuuQSB3oZPRHj3Q/800px-Isobaric_labeling.png","https://steemitimages.com/DQmcH3i1YRmZHo11QR4Ryf5LH8fuenYuVCiugMZebtzLGvs/tmt.png","https://steemitimages.com/DQmRFAoQGL2nFNBBqipZd1Bzm1bdmF9HuKGRiYWNmRHcoj2/andjelka.jpg"],"links":["https://en.wikipedia.org/wiki/File:Summary_of_RNA-Seq.svg","https://creativecommons.org/licenses/by/4.0/deed.en","http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004393","https://steemit.com/steemstem/@scienceangel/lab-diaries-2-making-my-own-transgenic-cell-line","https://en.wikipedia.org/wiki/File:Isobaric_labeling.png","https://creativecommons.org/licenses/by-sa/3.0/","https://www.nature.com/articles/nrg2934","https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2949280/","https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4863231/","http://www.rna-seqblog.com/rpkm-fpkm-and-tpm-clearly-explained/","https://www.ncbi.nlm.nih.gov/pubmed/28188531","https://en.wikipedia.org/wiki/Tandem_mass_tag","https://en.wikipedia.org/wiki/Isobaric_labeling","https://steemit.com/trending/steemstem","https://www.jason-brooks.com/","https://medicine.ekmd.huji.ac.il/En/Publications/ResearchersPages/pages/sharonaev.aspx"],"app":"steemit/0.1","format":"markdown"} |
| created | 2018-04-08 18:21:33 |
| last_update | 2018-04-15 12:28:18 |
| depth | 0 |
| children | 14 |
| last_payout | 2018-04-15 18:21:33 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 46.023 HBD |
| curator_payout_value | 13.346 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 8,509 |
| author_reputation | 113,585,841,123,698 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,010,490 |
| net_rshares | 13,170,102,544,669 |
| author_curate_reward | "" |
| voter | weight | wgt% | rshares | pct | time |
|---|---|---|---|---|---|
| pharesim | 0 | 78,062,214,431 | 0.09% | ||
| lafona-miner | 0 | 877,032,506,144 | 60% | ||
| kushed | 0 | 162,330,412,348 | 10.8% | ||
| bue | 0 | 13,987,719,597 | 100% | ||
| justtryme90 | 0 | 2,173,069,670,243 | 100% | ||
| anwenbaumeister | 0 | 271,593,944,455 | 10.8% | ||
| cmp2020 | 0 | 11,557,554,827 | 22% | ||
| lemouth | 0 | 158,913,348,484 | 40% | ||
| rjbauer85 | 0 | 3,224,198,868 | 60% | ||
| anarchyhasnogods | 0 | 93,373,002,541 | 30% | ||
| lamouthe | 0 | 13,693,011,728 | 60% | ||
| roy2016 | 0 | 62,638,320,373 | 12% | ||
| curie | 0 | 465,028,977,673 | 10.8% | ||
| cub1 | 0 | 39,273,376,603 | 22% | ||
| hendrikdegrote | 0 | 4,902,257,464,265 | 11.88% | ||
| steemstem | 0 | 1,485,324,638,831 | 60% | ||
| sethroot | 0 | 185,072,727 | 1.08% | ||
| brobear1995 | 0 | 203,107,003 | 5.4% | ||
| foundation | 0 | 5,300,591,699 | 60% | ||
| the-devil | 0 | 6,816,507,486 | 60% | ||
| thevenusproject | 0 | 30,236,851,615 | 60% | ||
| dna-replication | 0 | 15,378,452,181 | 60% | ||
| lenin-mccarthy | 0 | 340,755,631 | 5.4% | ||
| jgpro | 0 | 94,842,828 | 5.4% | ||
| heriafriadiaka | 0 | 144,754,992 | 10.8% | ||
| dyancuex | 0 | 140,456,410 | 5.4% | ||
| pacokam8 | 0 | 418,206,405 | 2.7% | ||
| borislavzlatanov | 0 | 4,673,014,404 | 60% | ||
| michelios | 0 | 6,220,900,423 | 2.16% | ||
| kirkins | 0 | 2,533,523,271 | 5% | ||
| awesomianist | 0 | 1,355,441,050 | 2.16% | ||
| jamhuery | 0 | 8,102,493,998 | 60% | ||
| jdc | 0 | 80,020,792 | 1.08% | ||
| devi1714 | 0 | 58,883,628 | 5.4% | ||
| jacalf | 0 | 84,283,198 | 10.8% | ||
| ninyea | 0 | 296,374,413 | 10.8% | ||
| kryzsec | 0 | 81,288,784,389 | 60% | ||
| markangeltrueman | 0 | 1,327,833,407 | 1.62% | ||
| odic3o1 | 0 | 133,320,563 | 6% | ||
| galberto | 0 | 187,817,332 | 0.32% | ||
| lrsm13 | 0 | 97,709,860 | 3.24% | ||
| tantawi | 0 | 981,165,732 | 10.8% | ||
| trumpman | 0 | 149,968,319,790 | 100% | ||
| steemanator | 0 | 53,563,950 | 10.8% | ||
| locikll | 0 | 4,436,201,417 | 21.6% | ||
| dber | 0 | 14,231,305,976 | 60% | ||
| mahdiyari | 0 | 2,338,735,365 | 3.24% | ||
| aboutyourbiz | 0 | 2,000,761,661 | 10.8% | ||
| kerriknox | 0 | 146,719,763,059 | 60% | ||
| howtostartablog | 0 | 436,014,509 | 0.54% | ||
| blessing97 | 0 | 2,164,788,547 | 60% | ||
| guchtere | 0 | 143,641,675 | 5.4% | ||
| suesa | 0 | 356,394,654,772 | 100% | ||
| cryptokrieg | 0 | 2,171,038,180 | 10.8% | ||
| zacherybinx | 0 | 81,736,358 | 10.8% | ||
| rockeynayak | 0 | 335,165,862 | 60% | ||
| ertwro | 0 | 22,502,693,358 | 60% | ||
| coloringiship | 0 | 165,000,137 | 0.54% | ||
| juanjdiaz89 | 0 | 2,166,435,177 | 60% | ||
| thinknzombie | 0 | 17,208,526,626 | 5.4% | ||
| nitesh9 | 0 | 13,622,334,610 | 60% | ||
| churchboy | 0 | 8,176,881,250 | 60% | ||
| tking77798 | 0 | 14,960,003,630 | 100% | ||
| jpederson96 | 0 | 58,831,185 | 1.5% | ||
| gambit.coin | 0 | 115,801,486 | 10.8% | ||
| mcw | 0 | 4,889,640,981 | 10% | ||
| himal | 0 | 4,201,422,501 | 60% | ||
| bachuslib | 0 | 18,215,556,078 | 100% | ||
| abigail-dantes | 0 | 934,335,745,058 | 60% | ||
| leczy | 0 | 3,556,104,251 | 60% | ||
| steemulator | 0 | 2,341,296,470 | 10.8% | ||
| phogyan | 0 | 325,793,354 | 5.4% | ||
| suravsingh | 0 | 684,984,032 | 60% | ||
| helgapn | 0 | 181,596,280 | 5.4% | ||
| krazypoet | 0 | 89,649,294 | 0.05% | ||
| joseg | 0 | 728,184,001 | 36% | ||
| kofspades | 0 | 75,181,113 | 5.4% | ||
| onethousandwords | 0 | 186,836,336 | 5.4% | ||
| akeelsingh | 0 | 2,014,295,828 | 60% | ||
| justdentist | 0 | 594,427,263 | 1% | ||
| muliadi | 0 | 92,525,029 | 5.4% | ||
| karyah1001 | 0 | 113,212,008 | 10.8% | ||
| purepinay | 0 | 27,807,568,799 | 3% | ||
| zest | 0 | 37,998,476,844 | 100% | ||
| felixrodriguez | 0 | 1,262,952,742 | 30% | ||
| dysprosium | 0 | 20,661,642,279 | 100% | ||
| pearlumie | 0 | 21,908,160,026 | 60% | ||
| gabox | 0 | 172,266,954 | 0.54% | ||
| intellectualsoul | 0 | 55,111,678 | 5.4% | ||
| runningman | 0 | 101,173,183 | 5.4% | ||
| infinitelearning | 0 | 272,042,682 | 5.4% | ||
| circleoffriends | 0 | 72,611,116 | 5.4% | ||
| bssman | 0 | 3,873,493,925 | 30% | ||
| simplifylife | 0 | 8,749,643,237 | 30% | ||
| rejzons | 0 | 56,098,743 | 1.08% | ||
| smafey | 0 | 128,903,234 | 5.4% | ||
| zanoni | 0 | 115,145,224 | 5.4% | ||
| marialefleitas | 0 | 119,543,775 | 5.4% | ||
| damzxyno | 0 | 64,469,440 | 3.24% | ||
| hillaryaa | 0 | 130,445,267 | 10.8% | ||
| jordanx2 | 0 | 534,622,200 | 5.4% | ||
| thatterrioguy | 0 | 339,128,303 | 8.1% | ||
| stbrians | 0 | 242,014,817 | 5.4% | ||
| mayowadavid | 0 | 4,037,274,626 | 30% | ||
| imamalkimas | 0 | 81,460,406 | 10.8% | ||
| dinglehopper | 0 | 81,662,127 | 5.4% | ||
| martis6 | 0 | 0 | 100% | ||
| peaceandwar | 0 | 1,295,765,290 | 5.4% | ||
| enzor | 0 | 918,504,588 | 30% | ||
| rmz | 0 | 63,286,574 | 5.4% | ||
| kieranpearson | 0 | 269,470,999 | 2.7% | ||
| bobdos | 0 | 535,578,050 | 1.08% | ||
| jesusj1 | 0 | 120,255,849 | 5.4% | ||
| carloserp-2000 | 0 | 9,332,178,326 | 60% | ||
| pangoli | 0 | 5,531,255,262 | 60% | ||
| rachelsmantra | 0 | 2,338,693,997 | 60% | ||
| altherion | 0 | 44,914,358,894 | 100% | ||
| christianschmidt | 0 | 1,279,234,198 | 10.8% | ||
| gra | 0 | 20,311,925,822 | 60% | ||
| artepoetico | 0 | 127,287,683 | 5.4% | ||
| wandersells | 0 | 57,418,295 | 5.4% | ||
| ifartrainbows | 0 | 335,353,292 | 15% | ||
| syalla | 0 | 343,683,445 | 60% | ||
| sci-guy | 0 | 139,946,628 | 60% | ||
| kerry234 | 0 | 125,070,616 | 10.8% | ||
| maidisangkot | 0 | 52,180,701 | 10.8% | ||
| katerinaramm | 0 | 19,702,613,585 | 46% | ||
| delph-in-holland | 0 | 116,015,875 | 5.4% | ||
| babangsunan | 0 | 107,682,248 | 100% | ||
| meetmysuperego | 0 | 271,560,136 | 2.7% | ||
| skycae | 0 | 1,931,340,467 | 10.8% | ||
| sireh | 0 | 1,293,025,845 | 2.7% | ||
| xanderslee | 0 | 683,361,081 | 10.8% | ||
| kenadis | 0 | 14,774,117,158 | 60% | ||
| amavi | 0 | 9,282,682,393 | 12% | ||
| enjoyy | 0 | 54,289,567 | 5.4% | ||
| robotics101 | 0 | 5,398,796,906 | 100% | ||
| tristan-muller | 0 | 483,144,953 | 60% | ||
| paulthebeloved | 0 | 67,221,085 | 5.4% | ||
| jaeydallah | 0 | 148,559,952 | 10.8% | ||
| hasan086 | 0 | 54,254,474 | 5.4% | ||
| alexs1320 | 0 | 36,966,713,555 | 100% | ||
| gentleshaid | 0 | 13,373,448,560 | 60% | ||
| zalandir | 0 | 885,742,566 | 5.4% | ||
| crescendoofpeace | 0 | 140,253,594 | 5.4% | ||
| tito36 | 0 | 64,286,663 | 10.8% | ||
| irza | 0 | 4,043,176,944 | 100% | ||
| sco | 0 | 32,596,463,215 | 100% | ||
| adetola | 0 | 2,124,257,326 | 60% | ||
| barut | 0 | 78,296,790 | 5.4% | ||
| evernew | 0 | 57,222,552 | 5.4% | ||
| dysfunctional | 0 | 2,492,319,400 | 30% | ||
| rasamuel | 0 | 123,567,187 | 5.4% | ||
| cordeta | 0 | 165,366,645 | 5.4% | ||
| cerventus | 0 | 80,408,459 | 5.4% | ||
| ariffashraff | 0 | 61,294,725 | 5.4% | ||
| speaklife | 0 | 108,878,701 | 10.8% | ||
| smanuels | 0 | 51,342,463 | 5.4% | ||
| bimijay | 0 | 61,482,069 | 10.8% | ||
| whileponderin | 0 | 1,247,314,508 | 60% | ||
| jlmol7 | 0 | 134,677,262 | 30% | ||
| mittymartz | 0 | 440,585,569 | 30% | ||
| bennettitalia | 0 | 182,970,892 | 0.54% | ||
| hadji | 0 | 2,650,009,717 | 60% | ||
| sakura1012 | 0 | 1,175,867,471 | 60% | ||
| scienceangel | 0 | 16,667,673,419 | 100% | ||
| adebayopaul | 0 | 60,171,370 | 10.8% | ||
| gwys | 0 | 78,124,143 | 5.4% | ||
| sikan-eyen | 0 | 67,350,606 | 10.8% | ||
| mrxplicit | 0 | 65,138,553 | 10.8% | ||
| acelad | 0 | 1,491,980,027 | 10.8% | ||
| fidelpoet | 0 | 435,383,497 | 10.8% | ||
| geekis | 0 | 50,601,052 | 10.8% | ||
| terrylovejoy | 0 | 3,672,337,425 | 12% | ||
| dynamicrypto | 0 | 150,530,164 | 1% | ||
| wisewoof | 0 | 1,208,251,931 | 5.4% | ||
| neneandy | 0 | 543,576,074 | 10.8% | ||
| strings | 0 | 119,792,881 | 5.4% | ||
| giddyupngo | 0 | 149,645,886 | 5.4% | ||
| steepup | 0 | 321,038,476 | 24% | ||
| zoricatech | 0 | 1,474,069,293 | 100% | ||
| aaronteng | 0 | 82,570,894 | 5.4% | ||
| debbietiyan | 0 | 219,152,093 | 5.4% | ||
| emirfirlar | 0 | 84,478,312 | 5.4% | ||
| rionpistorius | 0 | 1,184,631,237 | 30% | ||
| randomwanderings | 0 | 332,907,123 | 10.8% | ||
| heajin | 0 | 72,592,610 | 15% | ||
| deutsch-boost | 0 | 342,375,825 | 20% | ||
| wdoutjah | 0 | 455,836,320 | 5.4% | ||
| modernmclaire | 0 | 64,375,694 | 10.8% | ||
| jpmkikoy | 0 | 84,553,078 | 5.4% | ||
| caitycat | 0 | 170,214,326 | 5.4% | ||
| dexterdev | 0 | 11,103,055,265 | 60% | ||
| gio6 | 0 | 220,270,097 | 5.4% | ||
| loydjayme25 | 0 | 66,500,898 | 5.4% | ||
| ugonma | 0 | 789,716,171 | 60% | ||
| ajpacheco1610 | 0 | 357,611,846 | 30% | ||
| drkomoo | 0 | 673,075,664 | 60% | ||
| v1tko | 0 | 55,142,240 | 5.4% | ||
| alexdory | 0 | 6,408,924,454 | 60% | ||
| benleemusic | 0 | 2,654,887,489 | 0.54% | ||
| vinamra | 0 | 1,509,865,726 | 100% | ||
| kul0tzzz | 0 | 55,065,691 | 10.8% | ||
| chimtivers96 | 0 | 586,615,626 | 10.8% | ||
| jerscoguth | 0 | 67,595,010 | 10.8% | ||
| contentvoter | 0 | 751,342,562 | 50% | ||
| amirdesaingrafis | 0 | 72,123,210 | 5.4% | ||
| wrpx | 0 | 83,133,854 | 5.4% | ||
| shawnycx | 0 | 61,195,218 | 10.8% | ||
| anyes2013 | 0 | 286,726,436 | 30% | ||
| jaycem | 0 | 238,742,901 | 42% | ||
| pojgaerlan | 0 | 52,105,606 | 10.8% | ||
| positiveninja | 0 | 1,007,949,997 | 5.4% | ||
| the-doubled | 0 | 61,204,719 | 10.8% | ||
| e-troubled | 0 | 64,418,435 | 10.8% | ||
| theunlimited | 0 | 52,225,947 | 10% | ||
| cryptostyle | 0 | 149,865,202 | 30% | ||
| simplicitytech | 0 | 168,216,043 | 30% | ||
| wa2qr | 0 | 129,786,291 | 5.4% | ||
| pseudojew | 0 | 916,106,051 | 60% | ||
| yu-stem | 0 | 2,438,661,135 | 100% | ||
| effofex | 0 | 473,409,416 | 30% | ||
| wisata | 0 | 67,590,811 | 10.8% | ||
| de-stem | 0 | 22,088,325,511 | 60% | ||
| ikeror | 0 | 61,305,362 | 9.72% | ||
| fibrefox | 0 | 104,843,198 | 5.4% | ||
| ari16 | 0 | 171,415,130 | 30% | ||
| blockmountain | 0 | 159,661,277 | 3.24% | ||
| hatuvera | 0 | 203,991,050 | 10% | ||
| foniz | 0 | 63,816,109 | 5.4% | ||
| i-have-tested | 0 | 61,170,344 | 10.8% | ||
| aarontaggert | 0 | 61,938,830 | 10.8% | ||
| etaletai | 0 | 69,072,523 | 5.4% | ||
| tessaragabrielle | 0 | 110,886,937 | 10% | ||
| the-tourist | 0 | 64,624,948 | 10.8% | ||
| rafaljakobsen | 0 | 97,814,160 | 5.4% | ||
| snowyknight | 0 | 402,365,359 | 100% | ||
| trisolaran | 0 | 348,341,274 | 5.4% | ||
| educatie | 0 | 69,839,234 | 60% | ||
| niouton | 0 | 644,638,323 | 2.16% | ||
| flyyingkiwi | 0 | 592,937,148 | 100% | ||
| beautyinscience | 0 | 177,628,536 | 30% | ||
| star-vc | 0 | 336,472,090 | 60% | ||
| medicnet | 0 | 309,312,287 | 60% | ||
| communityisyou | 0 | 309,312,287 | 60% | ||
| parakazan | 0 | 82,027,754 | 10.8% | ||
| biomimi | 0 | 174,378,106 | 30% | ||
| mirzantorres | 0 | 55,063,280 | 10.8% | ||
| funster | 0 | 256,733,293 | 60% |
It looks amazing! I wish I would have more time to read more about this. I will probably be the first to use my brain as a storage device when it will be technically possible :D It's very interesting and I envy you for being able to do this, although I don't regret doing my job at all. Basically I would like to do everything! Could you clone me, pls?
| author | alexdory |
|---|---|
| permlink | re-scienceangel-lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms-20180412t083505780z |
| category | steemstem |
| json_metadata | {"tags":["steemstem"],"app":"steemit/0.1"} |
| created | 2018-04-12 08:35:03 |
| last_update | 2018-04-12 08:35:03 |
| depth | 1 |
| children | 1 |
| last_payout | 2018-04-19 08:35:03 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.051 HBD |
| curator_payout_value | 0.016 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 354 |
| author_reputation | 6,862,980,134,251 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,636,881 |
| net_rshares | 14,944,624,774 |
| author_curate_reward | "" |
| voter | weight | wgt% | rshares | pct | time |
|---|---|---|---|---|---|
| scienceangel | 0 | 14,944,624,774 | 100% |
I'm still practicing on cells (creating new species, etc.), but as soon as my skills are good enough you can be the first person to have the honor of being cloned by me :D
| author | scienceangel |
|---|---|
| permlink | re-alexdory-re-scienceangel-lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms-20180412t092701461z |
| category | steemstem |
| json_metadata | {"tags":["steemstem"],"app":"steemit/0.1"} |
| created | 2018-04-12 09:27:27 |
| last_update | 2018-04-12 09:27:51 |
| depth | 2 |
| children | 0 |
| last_payout | 2018-04-19 09:27:27 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.000 HBD |
| curator_payout_value | 0.000 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 171 |
| author_reputation | 113,585,841,123,698 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,643,443 |
| net_rshares | 0 |
Thank you for your post. :) I have voted for you: 🎁! To call me just write @contentvoter in a comment.
| author | contentvoter |
|---|---|
| permlink | re-lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms-20180408t182542 |
| category | steemstem |
| json_metadata | "" |
| created | 2018-04-08 18:25:57 |
| last_update | 2018-04-08 18:25:57 |
| depth | 1 |
| children | 0 |
| last_payout | 2018-04-15 18:25:57 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.068 HBD |
| curator_payout_value | 0.015 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 110 |
| author_reputation | 412,164,657,355 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,010,988 |
| net_rshares | 19,164,414,954 |
| author_curate_reward | "" |
| voter | weight | wgt% | rshares | pct | time |
|---|---|---|---|---|---|
| tallfishinthesea | 0 | 18,413,072,392 | 100% | ||
| contentvoter | 0 | 751,342,562 | 50% |
I'm curious if you're using other statistical tests for differential expression? Deseq2 comes to mind. I'm wondering that if you used that (or adjusted alpha for you p-value test multiple testing) if it might not cut down the initial number of genes and proteins to look at. I'm familiar with RNA-seq but have no proteomics experience. How big (in terms of memory) do those datasets get?
| author | effofex |
|---|---|
| permlink | re-scienceangel-lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms-20180409t193548882z |
| category | steemstem |
| json_metadata | {"tags":["steemstem"],"app":"steemit/0.1"} |
| created | 2018-04-09 19:35:48 |
| last_update | 2018-04-09 19:35:48 |
| depth | 1 |
| children | 3 |
| last_payout | 2018-04-16 19:35:48 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.212 HBD |
| curator_payout_value | 0.013 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 389 |
| author_reputation | 14,429,105,750,792 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,189,695 |
| net_rshares | 50,104,446,166 |
| author_curate_reward | "" |
| voter | weight | wgt% | rshares | pct | time |
|---|---|---|---|---|---|
| alexs1320 | 0 | 34,270,066,966 | 100% | ||
| scienceangel | 0 | 15,834,379,200 | 100% |
As I'm doing the analyses in biology and chemistry I understand your standing point. In almost all the cases, we are able to offer them "better" math, but... Those things are alive. We often start with bad data or we get qualitatively the same thing at the end. I also noticed that sometimes we have "one-time hit" after application of exotic math that works well, but only once, on one dataset. Due to this, if the procedure works for the data (signals) - don't touch it, you will break something :) If they are stuck with data we are coming to the rescue.
| author | alexs1320 |
|---|---|
| permlink | re-effofex-re-scienceangel-lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms-20180409t195137160z |
| category | steemstem |
| json_metadata | {"tags":["steemstem"],"app":"steemit/0.1"} |
| created | 2018-04-09 19:51:39 |
| last_update | 2018-04-09 19:51:39 |
| depth | 2 |
| children | 0 |
| last_payout | 2018-04-16 19:51:39 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.000 HBD |
| curator_payout_value | 0.000 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 561 |
| author_reputation | 150,945,165,388,638 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,191,634 |
| net_rshares | 1,831,297,464 |
| author_curate_reward | "" |
| voter | weight | wgt% | rshares | pct | time |
|---|---|---|---|---|---|
| effofex | 0 | 1,831,297,464 | 100% |
Thank you for your comment! Basically, DESeq2 would not improve analysis from the biological point of view that much, improvement may be visible in those samples that have large deviation caused by measuring error only... To obtain biological meaning of datasets, I use GSEA which is based on Kolmogorov–Smirnov algorithm, and then I analyse biological pathways and genes within them. For proteomics data, size is not even close as of RNA-seq - you get your results in one Excel file :)
| author | scienceangel |
|---|---|
| permlink | re-effofex-re-scienceangel-lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms-20180409t195007329z |
| category | steemstem |
| json_metadata | {"tags":["steemstem"],"app":"steemit/0.1"} |
| created | 2018-04-09 19:50:09 |
| last_update | 2018-04-09 19:50:09 |
| depth | 2 |
| children | 1 |
| last_payout | 2018-04-16 19:50:09 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.000 HBD |
| curator_payout_value | 0.000 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 486 |
| author_reputation | 113,585,841,123,698 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,191,457 |
| net_rshares | 1,872,917,861 |
| author_curate_reward | "" |
| voter | weight | wgt% | rshares | pct | time |
|---|---|---|---|---|---|
| effofex | 0 | 1,872,917,861 | 100% |
That makes sense. I arrived at DESeq2 through examining microbial population changes and it's been super helpful in that context. It's always fun to see how a tool of choice can vary in efficacy between problems.
| author | effofex |
|---|---|
| permlink | re-scienceangel-re-effofex-re-scienceangel-lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms-20180409t195513128z |
| category | steemstem |
| json_metadata | {"tags":["steemstem"],"app":"steemit/0.1"} |
| created | 2018-04-09 19:55:09 |
| last_update | 2018-04-09 19:55:33 |
| depth | 3 |
| children | 0 |
| last_payout | 2018-04-16 19:55:09 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.000 HBD |
| curator_payout_value | 0.000 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 212 |
| author_reputation | 14,429,105,750,792 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,192,047 |
| net_rshares | 0 |
This is quite a post... I really didn't understand much on this topic. Your post was helpful great work
| author | joeycrack |
|---|---|
| permlink | re-scienceangel-lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms-20180409t200328615z |
| category | steemstem |
| json_metadata | {"tags":["steemstem"],"app":"steemit/0.1"} |
| created | 2018-04-09 19:52:54 |
| last_update | 2018-04-09 19:52:54 |
| depth | 1 |
| children | 0 |
| last_payout | 2018-04-16 19:52:54 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.000 HBD |
| curator_payout_value | 0.000 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 103 |
| author_reputation | 1,594,382,480,782 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,191,776 |
| net_rshares | 0 |
Hey for the people who din't listen to their 8-grade teachers # BASICALLY! There are three basic types of RNA used for protein synthesis of the body Which are mRNA/tRNA/ and rRNA So it basically follows this way # DNA > mRNA > tRNA > rRNA > Protein
| author | kettleyer-chan |
|---|---|
| permlink | re-scienceangel-lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms-20180409t064055828z |
| category | steemstem |
| json_metadata | {"tags":["steemstem"],"app":"steemit/0.1"} |
| created | 2018-04-09 06:40:54 |
| last_update | 2018-04-09 06:40:54 |
| depth | 1 |
| children | 2 |
| last_payout | 2018-04-16 06:40:54 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.000 HBD |
| curator_payout_value | 0.000 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 250 |
| author_reputation | -9,019,054,113 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,086,705 |
| net_rshares | 0 |
Lol quite funny... Nice breakdown
| author | joeycrack |
|---|---|
| permlink | re-kettleyer-chan-re-scienceangel-lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms-20180409t200059019z |
| category | steemstem |
| json_metadata | {"tags":["steemstem"],"app":"steemit/0.1"} |
| created | 2018-04-09 19:50:33 |
| last_update | 2018-04-09 19:50:33 |
| depth | 2 |
| children | 1 |
| last_payout | 2018-04-16 19:50:33 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.000 HBD |
| curator_payout_value | 0.000 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 33 |
| author_reputation | 1,594,382,480,782 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,191,494 |
| net_rshares | 0 |
heheheh
| author | kettleyer-chan |
|---|---|
| permlink | re-joeycrack-re-kettleyer-chan-re-scienceangel-lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms-20180410t001046320z |
| category | steemstem |
| json_metadata | {"tags":["steemstem"],"app":"steemit/0.1"} |
| created | 2018-04-10 00:10:48 |
| last_update | 2018-04-10 00:10:48 |
| depth | 3 |
| children | 0 |
| last_payout | 2018-04-17 00:10:48 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.000 HBD |
| curator_payout_value | 0.000 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 7 |
| author_reputation | -9,019,054,113 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,218,297 |
| net_rshares | 0 |
Congratulations @scienceangel! You have completed some achievement on Steemit and have been rewarded with new badge(s) : [](http://steemitboard.com/@scienceangel) Award for the number of upvotes Click on any badge to view your own Board of Honor on SteemitBoard. For more information about SteemitBoard, click [here](https://steemit.com/@steemitboard) If you no longer want to receive notifications, reply to this comment with the word `STOP` > Upvote this notification to help all Steemit users. Learn why [here](https://steemit.com/steemitboard/@steemitboard/http-i-cubeupload-com-7ciqeo-png)!
| author | steemitboard |
|---|---|
| permlink | steemitboard-notify-scienceangel-20180414t214544000z |
| category | steemstem |
| json_metadata | {"image":["https://steemitboard.com/img/notifications.png"]} |
| created | 2018-04-14 21:45:42 |
| last_update | 2018-04-14 21:45:42 |
| depth | 1 |
| children | 0 |
| last_payout | 2018-04-21 21:45:42 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.078 HBD |
| curator_payout_value | 0.000 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 683 |
| author_reputation | 38,975,615,169,260 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 50,095,376 |
| net_rshares | 15,555,602,492 |
| author_curate_reward | "" |
| voter | weight | wgt% | rshares | pct | time |
|---|---|---|---|---|---|
| scienceangel | 0 | 15,555,602,492 | 100% |
Very informative and helpful.Thanks for this post and keep it up.
| author | tanmoy016 |
|---|---|
| permlink | re-scienceangel-lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms-20180409t063115505z |
| category | steemstem |
| json_metadata | {"tags":["steemstem"],"app":"steemit/0.1"} |
| created | 2018-04-09 06:31:18 |
| last_update | 2018-04-09 06:31:18 |
| depth | 1 |
| children | 0 |
| last_payout | 2018-04-16 06:31:18 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.000 HBD |
| curator_payout_value | 0.000 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 65 |
| author_reputation | -716,823,670,032 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,085,612 |
| net_rshares | 0 |
Processing massive data sets like the ones you've generated are some of the biggest challenges in modern molecular biology. Looking forward to reading how you process this data.
| author | tking77798 |
|---|---|
| permlink | re-scienceangel-lab-diaries-4-whole-transcriptome-and-proteome-analysis-by-rna-sequencing-and-tmt-ms-20180409t121812921z |
| category | steemstem |
| json_metadata | {"tags":["steemstem"],"app":"steemit/0.1"} |
| created | 2018-04-09 12:18:15 |
| last_update | 2018-04-09 12:18:15 |
| depth | 1 |
| children | 0 |
| last_payout | 2018-04-16 12:18:15 |
| cashout_time | 1969-12-31 23:59:59 |
| total_payout_value | 0.108 HBD |
| curator_payout_value | 0.029 HBD |
| pending_payout_value | 0.000 HBD |
| promoted | 0.000 HBD |
| body_length | 178 |
| author_reputation | 14,484,662,307,666 |
| root_title | "Lab Diaries #4 - Whole Transcriptome and Proteome Analysis by RNA Sequencing and TMT-MS" |
| beneficiaries | [] |
| max_accepted_payout | 1,000,000.000 HBD |
| percent_hbd | 10,000 |
| post_id | 49,126,082 |
| net_rshares | 30,743,206,754 |
| author_curate_reward | "" |
| voter | weight | wgt% | rshares | pct | time |
|---|---|---|---|---|---|
| tking77798 | 0 | 14,239,111,417 | 100% | ||
| scienceangel | 0 | 16,504,095,337 | 100% |
 hiveblocks
hiveblocks